Deep learning tools in PD 3.0 (CHIMERYS and INFERYS) for TMT analysis
31:42
Deep learning tools in PD 3.0 (CHIMERYS and INFERYS) for TMT analysis
Related Videos
In Mass Spectrometry Videos
-
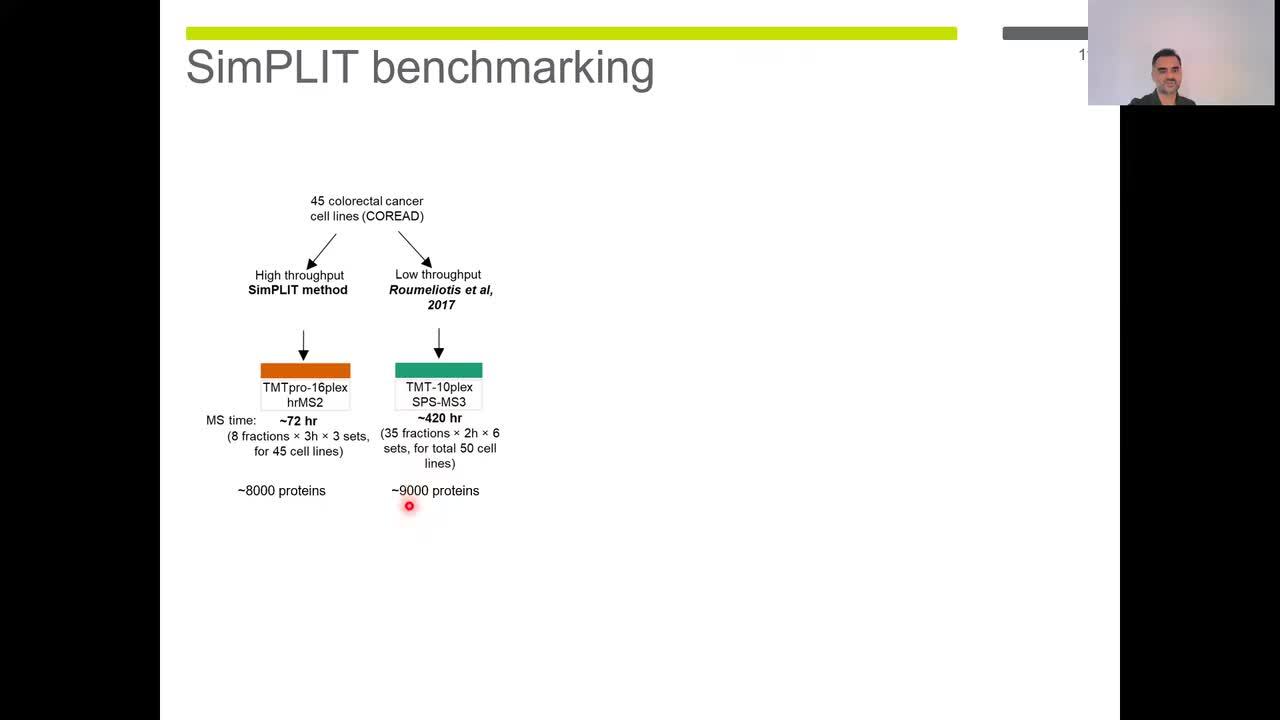
Play video SimPLIT: Simplified sample preparation for large-scale isobaric tagging proteomics
SimPLIT: Simplified sample preparation for large-scale isobaric tagging proteomics
A simplified and cost effective one-pot reaction workflow in a 96-well plate format (SimPLIT), based on a sodium deoxycholate lysis buffer, that minimizes processing steps and demonstrates improved reproducibility compared to alternative approaches.
20:18
-
Play video Measuring Protein-Turnover with Heavy Isotope Labeling and Complement Reporter Ions
Measuring Protein-Turnover with Heavy Isotope Labeling and Complement Reporter Ions
Measuring Protein-Turnover with Heavy Isotope Labeling and Complement Reporter Ions
29:34
-
Play video Orbitrap Astral MS Cinematic video
Orbitrap Astral MS Cinematic video
The Thermo Scientific Orbitrap Astral Mass Spectrometer enables you to dramatically expand the scale and scope of your experiments.
1:09
-
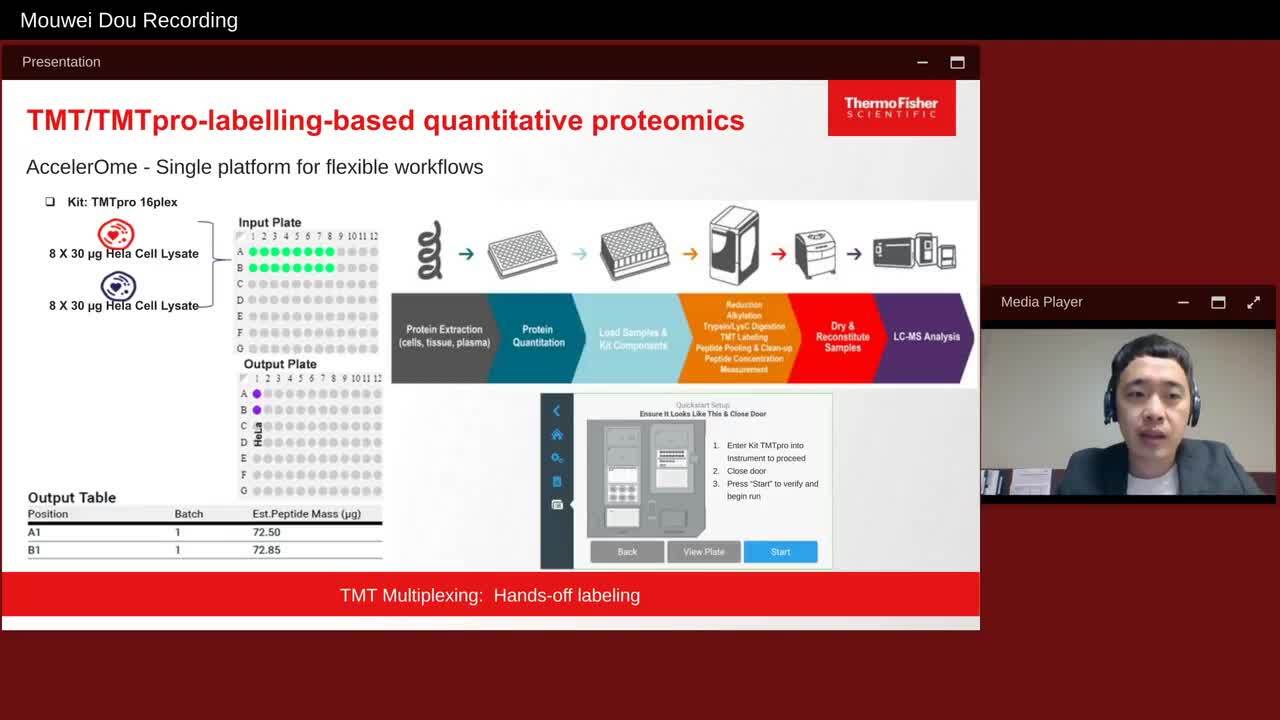
Play video An Automated Sample Preparation Solution for Mass Spectrometry-based Proteomics
An Automated Sample Preparation Solution for Mass Spectrometry-based Proteomics
The automated platform enables hands-off protein lysis, DNA removal, protein reduction and alkylation, digestion, TMT labeling, pooling and cleanup, and peptide concentration measurement, providing several workflows and companion reagents to perform
18:25
-
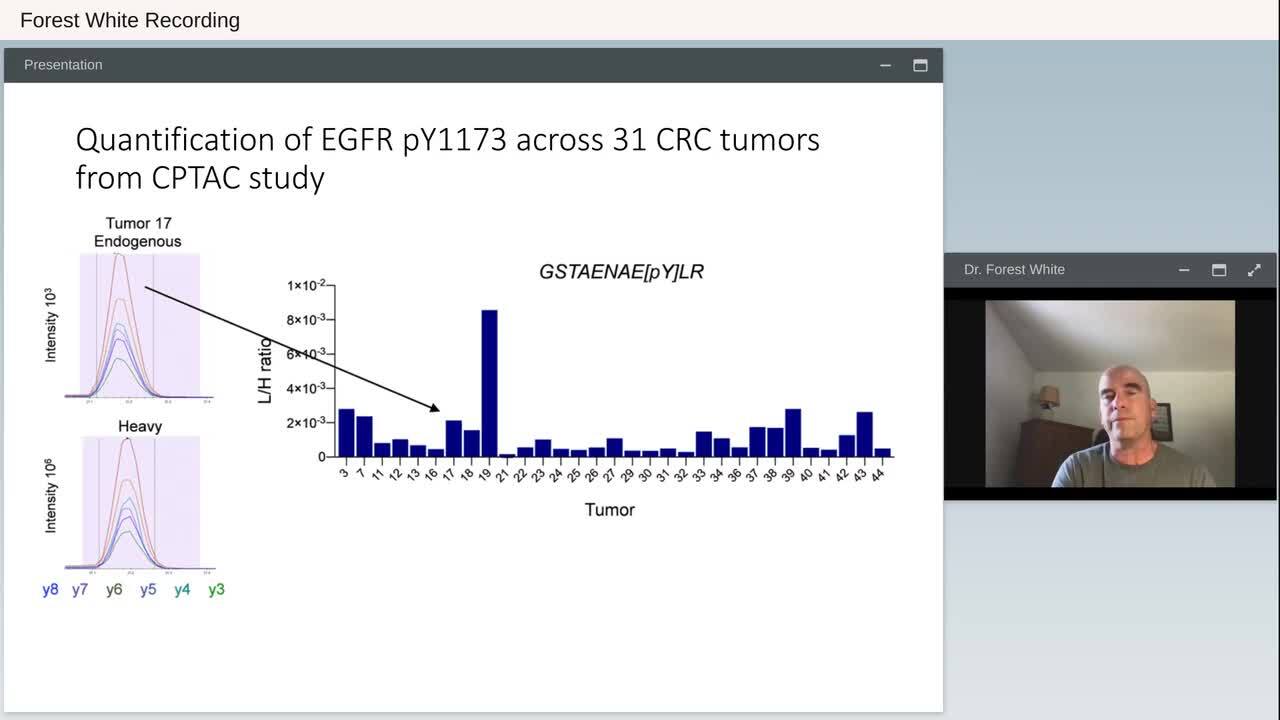
Play video Clinical Phosphoproteomics to Uncover New Therapeutic Strategies
Clinical Phosphoproteomics to Uncover New Therapeutic Strategies
Clinical Phosphoproteomics to Uncover New Therapeutic Strategies
26:22
-
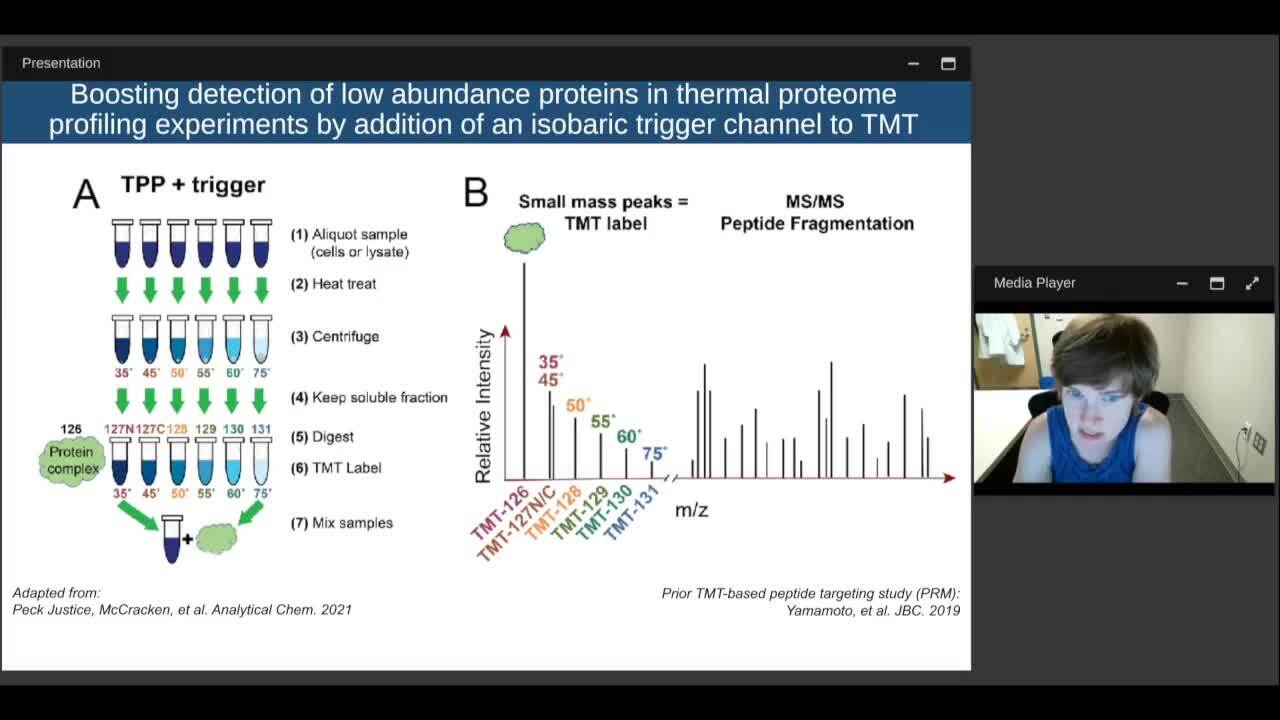
Play video Thermal Proteome Profiling for Functional and Molecular Characterization of Variant Proteins
Thermal Proteome Profiling for Functional and Molecular Characterization of Variant Proteins
We propose the use of thermal proteome profiling (TPP) for studying the impact of these disease related sequence variants on a systems wide level. TPP is a bottom up, TMT-based, quantitative mass spectrometry technique for determining the melt curves
30:52